Bioinformatics & Genomic Data Analysis
Using analysis pipelines developed specifically for variant detection, sequence data is aligned to available reference sequences. SNPs, indels (insertions and deletions), and structural variants are detected, quality-filtered, and annotated (coding, non-coding, synonymous, non-synonymous, etc.). In order to identify novel or rare variants, the variants are compared to an in-house database of known variants that includes the latest dbSNP and 1000Genomes data as well as other known variants from multiple organisms and publicly available data sets.
Transcriptome (RNA-Seq) data can be analyzed to determine gene or isoform level expression profiles, sequence variation, and differential expression between multiple conditions and/or timepoints. Included in this pipeline is the alignment of reads to a reference genome, expression analysis, differential expression analysis, isoform analysis, and differential isoform analysis. Results are output as spreadsheets containing statistics, differentially expressed genes, isoforms and differentially expressed isoforms as well as pdf plots and figures such as heat maps and principle component analyses.
We process amplicon sequence data from 16S, 18S, ITS, and custom amplicon projects using an in-house informatics workflow. This is offered primarily as a package in combination with our microbiome library and sequencing services but can also be performed on pre-existing data sets. Included in the analysis pipeline are the production of QC reports, taxonomic assignments, and assorted analysis (heatmaps, diversity measures) according to the project needs. Additional statistical analysis is offered to accommodate needs for advanced analysis relating to modeling and in-depth analysis.
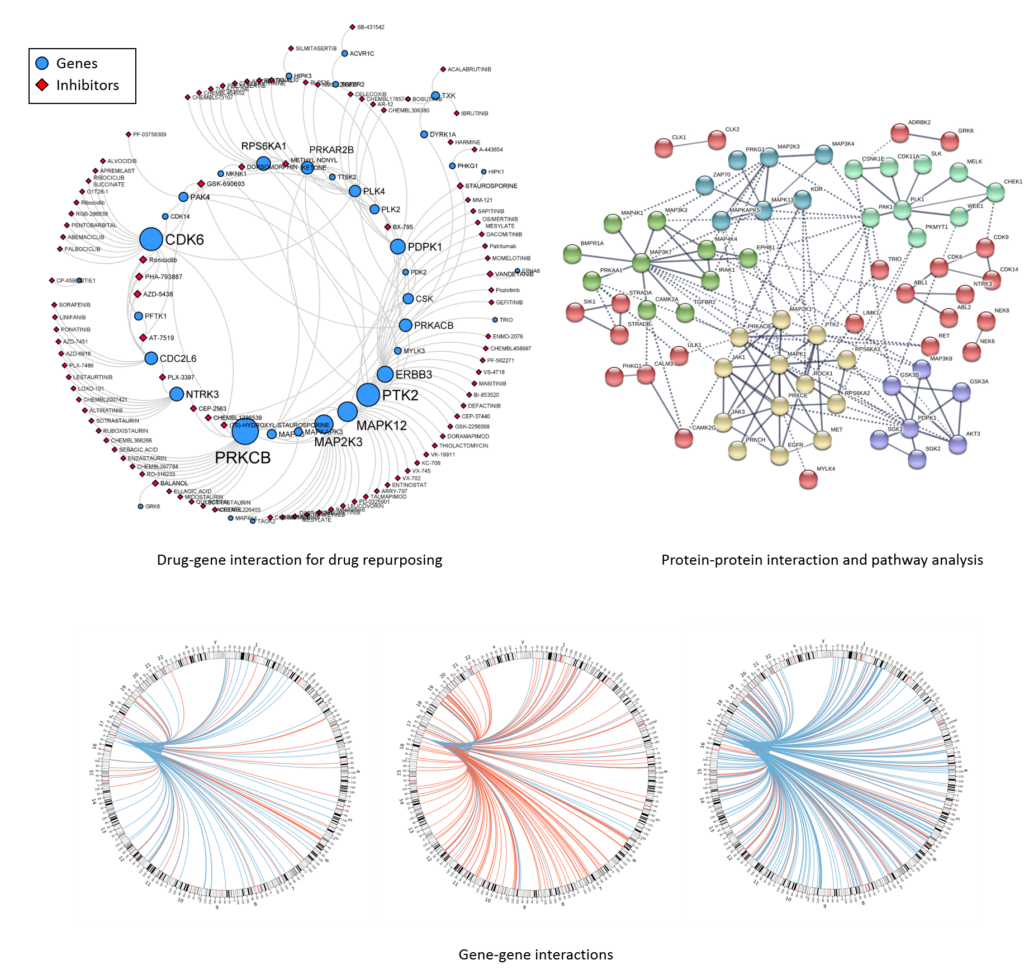
Given a set of differentially regulated gene set associated with a particular phenotype, we use software packages, developed specifically to analyze gene pathways and networks to find associations with functional profiles, tissue or disease specific biomarkers, and other genes in the same pathways and networks.
Our computational infrastructure and expertise enable cutting-edge custom analysis for a broad range of omics applications. We will customize an analysis plan for each project in close consultation with each investigator. Please request a consultation to discuss custom analysis projects.