COVID-19 Variant Detection and Sequencing
APPLICATIONS
Can this test be used for diagnosis of patients suspected of novel coronavirus?
This test has been authorized by FDA under an emergency use authorization (EUA) for use as an in vitro diagnostic test for the detection and/or diagnosis of COVID-19.
Can this test be used for surveillance?
COVIDSeq can be used to perform strain typing for monitoring virus evolution and epidemiology. It can be used to report virus sequence in a clinical sample for Public Health and research applications.
OVERVIEW
The Illumina COVIDSeq Test is intended for detection of SARS-CoV-2 virus RNA and virus genome analysis. Insight into the SARS-CoV-2 strain present in the sample enables tracking of virus strains. This amplicon-based NGS test includes 2019-nCoV primers designed to detect RNA from the SARS-CoV-2 virus in nasopharyngeal, oropharyngeal, and mid-turbinate nasal swabs from patients with signs and symptoms of infection who are suspected of COVID-19.
KEY FEATURES AND BENEFITS
- Accurate: Detects 98 targets on SARS-CoV-2 for highly accurate detection
- Comprehensive: Detects SARS-CoV-2 virus RNA; reports consensus genome calling for virus analysis under research use
- Quality Controlled: Built in quality control features in every reaction
- Flexible: Seamless end-to-end workflow.
COMPREHENSIVE COVID-19 SEQUENCING REPORTS
- Full COVID-19 strain variant identification
- Analysis of identified SARS-CoV-2 strain lineage and mutations
- Genome coverage, SNP, Consensus Genome, and raw sequencing analysis review
RAPID AND COST EFFECTIVE
(>3000 Results in 12 Hours)
At AGTC Genomics, up to 1536 to 3072 results can be processed on the NovaSeq 6000 system in 12 hours, coupled with the DRAGEN COVIDSeq Test Pipeline for rapid analysis.
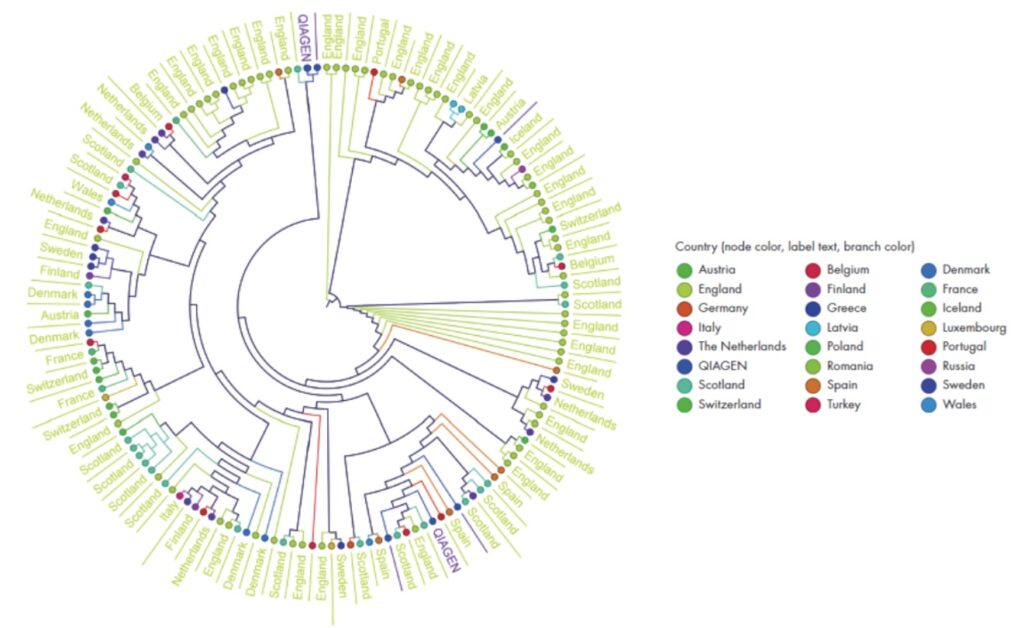
An example circular cladogram. In your report, it will show hypothetical phylogenetic relationships among the samples.

