De novo Sequencing
- Virulence research
- Drug resistance mechanism
- Vaccine development
- Epidemiology
- Microbial evolution
OVERVIEW
De novo sequencing can sequence the genome of a species without any reference genome information, splice and assemble it by bioinformatics analysis methods, and obtain the genome sequence map of the species, to promote the follow-up research of the species. It offers reference genome assembly for rarely studied species. Using de novo sequencing, researchers can obtain the genomic information of microbes providing a fresh start for exploring the genetic structure and functions, studying the evolutionary origin of microbial populations, as well as developing potential applications of these abundant microbes in medicine, disease, agriculture, and the environment. De novo sequencing can also be applied to research of animals, and plants, including phylogenetic studies, analysis of species diversity, genetic markers, and other genomic research.
INPUT REQUIREMENTS
- ≥ 200 ng of purified genomic DNA
- A260/280 = 1.8-2.0
- No degradation, No contamination
What’s Included
- Sample Receipt and Initial QC
- Library Construction and QC
- Sequencing and Data Delivery
- Bioinformatics Analysis
Data Deliverable
- Bacterial and Fungal Draft Map
- Genome preliminarily assembly
- Genome component analysis
- Gene function annotation
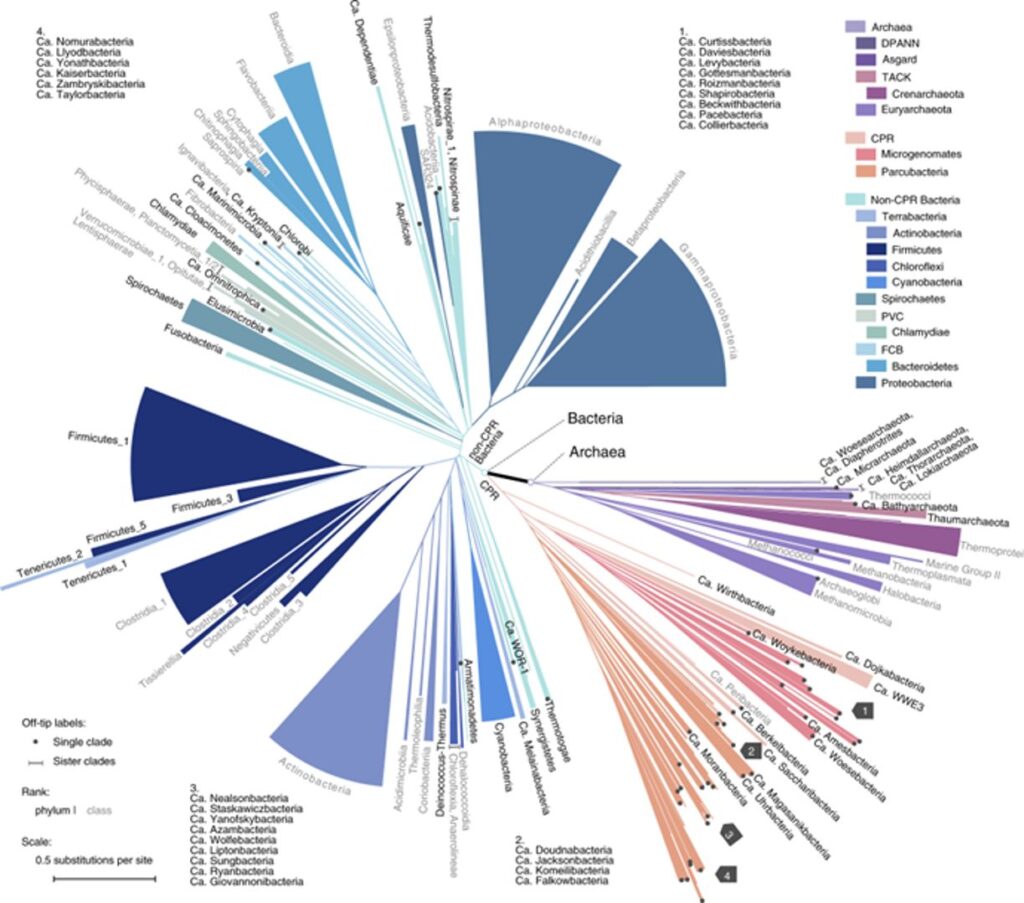
Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea.
Nat Commun 2019, 10(1):5477.

