A High-resolution View Of The Entire Genome
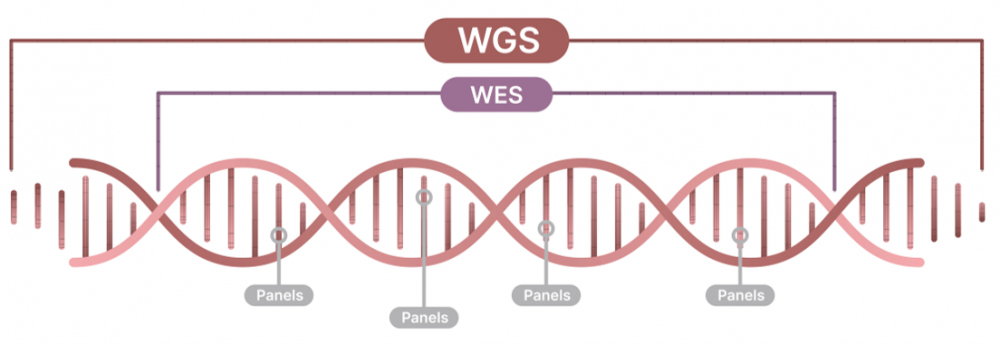
Whole genome sequencing (WGS) identifies almost all changes in a patient’s DNA by sequencing both the entire protein coding and the non-coding regions of the genome.
Today there are millions of patients suffering from misdiagnosed or undiagnosed genetic diseases as a result of insufficient genetic testing. Although in certain cases approaches like single gene testing, panel testing, or microarrays can identify the cause of a disease, these analyses are ultimately limited and can fail to reveal the full genetic cause. WGS, in contrast, overcomes such limitations and is the only test that can detect nearly all types of disease-causing genetic variants in one single test1,2.
Most studies on genetic diseases have been heavily biased towards variants in gene coding regions, but this only accounts for approx. 1-2% of a patient’s entire genome. Recently, however, a growing body of studies have demonstrated that clinical WGS offers a more comprehensive analysis than whole exome sequencing (WES) and can provide molecular diagnosis where WES can not1-5. Non-coding variants, such as intergenic and intronic pathogenic variants, are growing in number and importance, spanning from sequence variants to more complex structural variations5,6.
When is WGS Recommended?
- Preventive cares. Genetic risk assessment and screening for healthy individual in order to predict and potentially prevent the onset of diseases.
- Family planning & carrier screen. An accurate and safe DNA testing to check if you are a carrier for certain genetic conditions and understand if you have a higher chance to have a child with that condition.
- Complex medical problems and rare diseases. Patients with symptoms that are very broad, complex, or unspecific, not pointing towards a specific disease or typical phenotype, or patients with a long list of inconclusive prior differential diagnoses, or who have exhausted other testing options.
- When a rapid diagnosis is a medical necessity. When there is no time for serial testing strategies, a rapid diagnosis can be critical for timely clinical decision-making in critically ill newborn infants and children in the Neonatal Intensive Care Units (NICU) or Paediatric Intensive Care Units (PICU).


Features & Performance
| Highly uniform genome and mitochondrial genome coverage |
|
| Advanced detection of nearly all types of variants in one single test | Highly sensitive and specific detection of SNVs, InDels, SVs including CNVs, and mtDNA variants with heteroplasmy ≥15%
|
SNVs: single nucleotide variants; InDels: small insertions/deletions; CNVs: copy number variations; SVs: structural variants.
Sample Preparation Instructions
All clinical materials should be collected with approved methods to avoid contamination and cross contamination. A sterile environment must be maintained when collecting samples. Our sample preparation requirements are detailed in the table below:
| EDTA Blood | ≥ 1 mL |
| Buccal swab | 1 Oragene™ OCR-100 buccal swab collection set |
| Saliva | 1 Oragene™ OG-610 saliva collection set |
| Ready to use DNA | ≥ 1 µg For DNA samples, we recommend extraction using Qiagen kits, elution in water in 20 µL volume (min), plus evaluation of 260/280 Ratio (1.6 - 2.1) and fluorescent dye quantification of concentration (> 10 ng/µL) before sending. |
Sample Transport
EDTA Blood, buccal swabs, saliva, and DNA samples can be sent via regular mail at room temperature. Liquid blood is stable for up to 4 days during transport.
Shipping Instructions
Sample should be sent to:
AGTC Genomics Sdn Bhd (1428365-D)
J2-1, Pusat Perdagangan Bukit Jalil,
Jalan Persiaran Jalil 1,
Bukit Jalil,
57000 Kuala Lumpur,
Malaysia
References
1. Lionel et al. 2018, PMID: 28771251
2. Sanghvi et al. 2018, PMID: 9144510
3. Clark et al. 2018; PMID: 30002876
4. Stavropoulos et al. 2016, PMID: 28567303
5. Bertoli-Avella et al. 2020, PMID: 32860008
6. Bick et al. 2019, PMID: 31023718

