Whole Transcriptomic Sequencing (WTS)
- Determining differential gene expression, single nucleotide variations, and fusion gene in single run.
- Exploring miRNA sponge and target regulatory elements.
- Investigating regulatory networks among lncRNA/circRNA-miRNA-gene pairs.
OVERVIEW
Whole Transcriptome Sequencing (WTS) allows for the characterization of all types of RNA transcripts (coding and non-coding RNAs) of a particular organism, irrespective of whether or not they are polyadenylated. Whole transcriptome sequencing is advocated for both differential expression analysis as well as discovery work.
Our Whole Transcriptome Sequencing service offer a range of solutions tailored to your study’s needs. Our service is optimized to yield the highest quality, most reliable results, from differential expression analysis to the discovery and identification of mutations and fusion transcripts, even in FFPE tissue.
INPUT REQUIREMENTS
- 250ng of purified total RNA
- FFPE* tissue, fresh/frozen tissue, blood, or cell pellets
* Due to the inherent nature of samples derived from FFPE to contain degraded and crosslinked nucleic acids, all samples submitted are accepted “on risk” and subject to billing regardless of data quality.
What’s Included
- Sample Receipt and Initial QC
- Library Construction and QC
- Sequencing and Data Delivery
- Bioinformatics Analysis
Data Deliverable
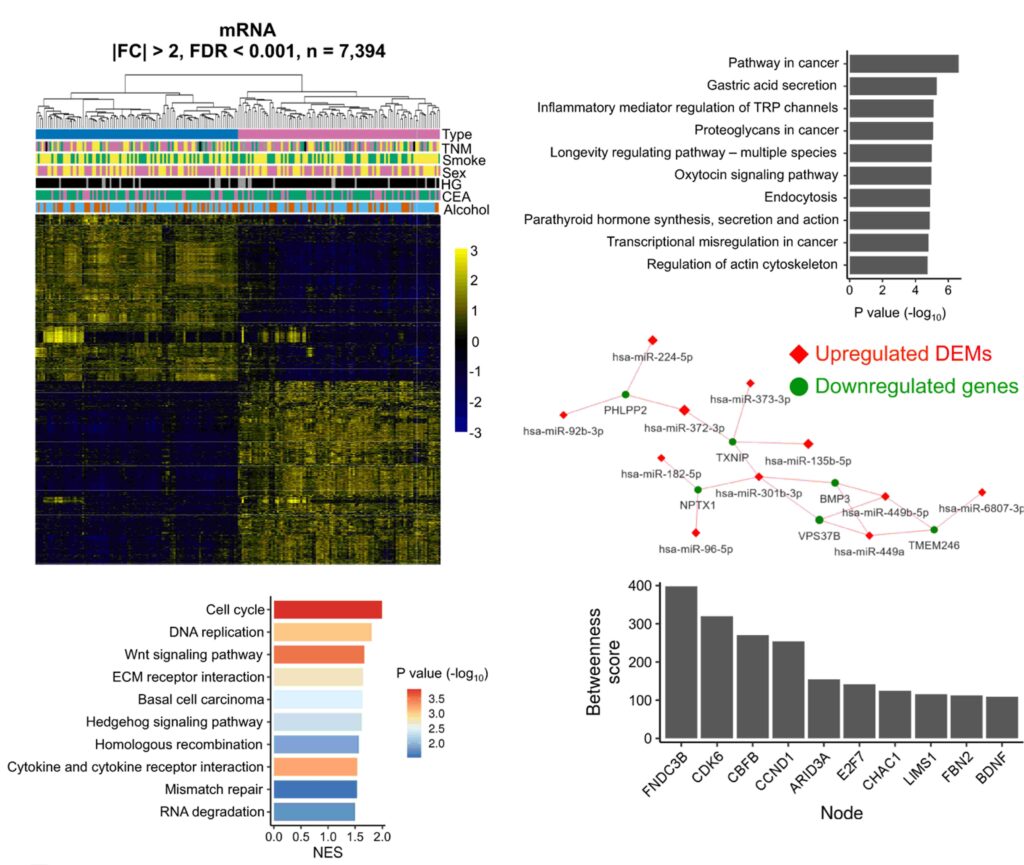
- Quantitative expression profiles
- Alternative splicing analysis
- Fusion gene analysis
- Time series analysis
- Gene ontology analysis
- Pathway enrichment analysis
- Hierarchical clustering analysis
- Protein-Protein Interaction (PPI) analysis