OncoDx™
Comprehensive Gene Profiling (CGP)
Personalized Gene Profiling For Precision Cancer Care
What if
we can give the right therapy for the right patient
Bring cancer into focus with comprehensive genomic profiling
OncoDx™ Comprehensive Gene Profiling provides you with a comprehensive analysis of your cancer’s genetic makeup, giving you and your healthcare team the information you need to make informed decisions about your treatment options. By looking at all the genes that may be involved in your cancer, we can get a deeper understanding of the underlying causes and develop more effective, targeted treatments.
With this information, you can benefit from personalized medicine, where treatments are selected based on the specific genetic mutations present in your cancer cells. This approach has been shown to be more effective than traditional, one-size-fits-all treatments, and can lead to better outcomes and improved quality of life.
Don’t settle for a limited understanding of your cancer. Get the full picture with our comprehensive gene profiling service and take control of your health today.
Key Benefits of CGP:
Detect Multiple Biomarkers in a Single Assay
CGP can detect biomarkers at nucleotide-level resolution and typically comprises all major genomic variant classes (SNVs, indels, CNVs, fusions, splice variants), as well as large genomic signatures (TMB, MSI), maximizing the ability to find clinically actionable alterations.

Consolidate Testing to Save Time and Precious Samples
CGP consolidates biomarker detection into a single multiplex assay, eliminating the need for sequential testing. With a single test, you can assess the most prevalent as well as rare biomarkers. By assessing all biomarkers at once, you may increase the chances of finding an actionable alteration. This potentially provides faster results, limits the input of precious biopsy samples, and may reduce the risks and cost associated with rebiopsy.

Identify Actionable Alterations
CGP can offer both actionable and potentially actionable results to help identify more effective therapeutic paths and innovative clinical trial options for cancer patients. When tissue biopsies are unavailable, CGP from a liquid biopsy may provide helpful information about a tumor’s genomic makeup. CGP using tissue and liquid biopsy together may reveal more insights into a tumor’s composition.
Continuously Evolving
Comprehensive genomic profiling can help you keep up to date with the most recent treatment options available throughout the patient’s treatment journey.
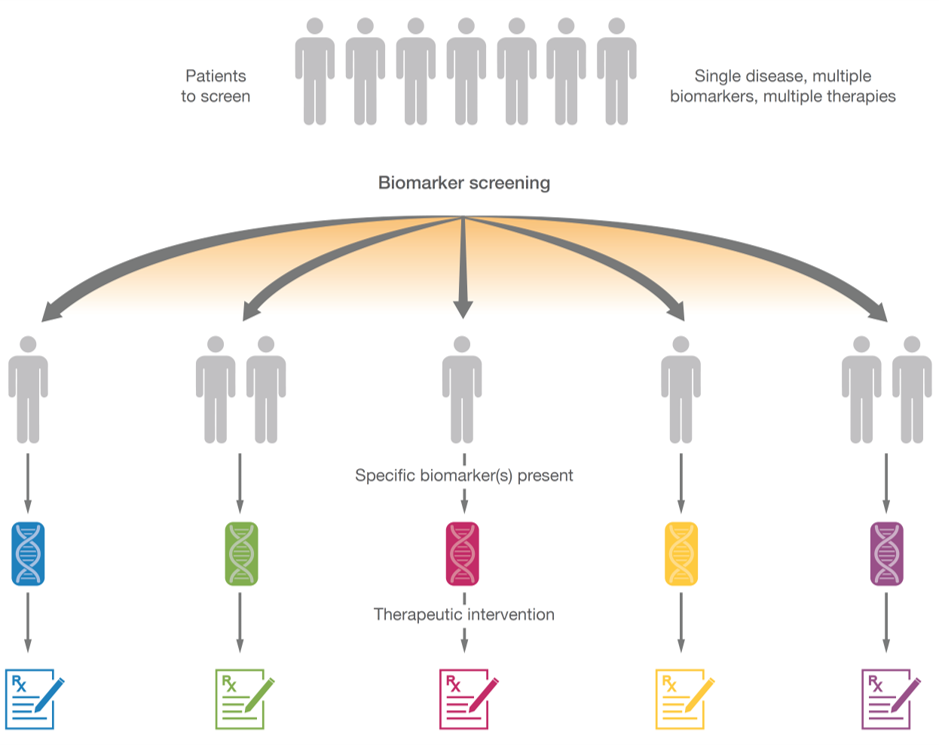
The right therapy for the right patient
Molecular matching of a patient’s cancer with an approved or investigational therapy is essential for increasing the chances of achieving better clinical outcomes. Multiple studies have shown that genomically matching patients to targeted therapies or immunotherapies results in improved clinical outcomes. Additionally, CGP can improve patient enrollment in genomically matched clinical trials.
Matched therapy can lead to better patient outcomes. 8-11

How to Order
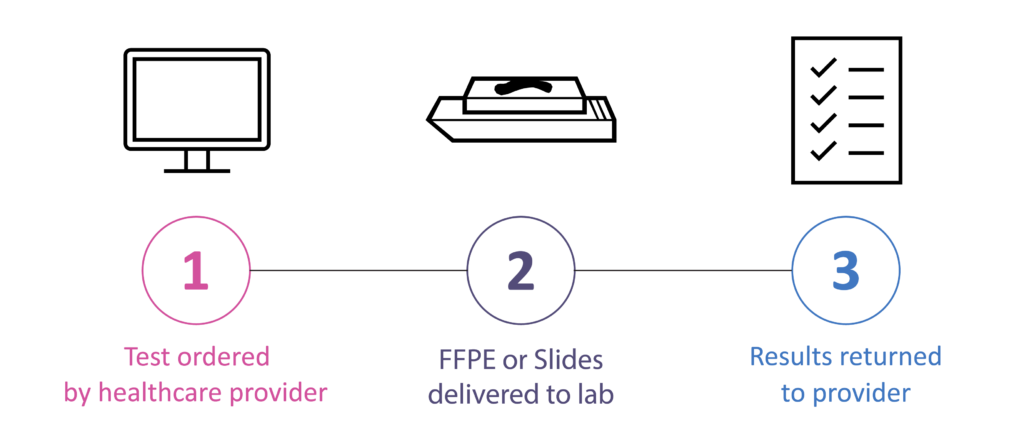
Sample Preparation Instructions
All clinical materials should be collected with approved methods to avoid contamination and cross contamination. A sterile environment must be maintained when collecting samples. Our sample preparation requirements are detailed in the table below:
| FFPE Tissue | 4 sections, each with a thickness of 10 µm |
| Ready to use DNA and RNA | ≥ 40 ng of DNA ≥ 40 ng of RNA |
Sample Transport
FFPE and DNA/RNA samples can be sent via regular mail at room temperature.
Shipping Instructions
Sample should be sent to:
AGTC Genomics Sdn Bhd (1428365-D)
J2-1, Pusat Perdagangan Bukit Jalil,
Jalan Persiaran Jalil 1,
Bukit Jalil,
57000 Kuala Lumpur,
Malaysia
References
1. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34.
2. Pestinger V, Smith M, Sillo T, et al. Use of an Integrated Pan-Cancer Oncology Enrichment Next-Generation Sequencing Assay to Measure Tumour Mutational Burden and Detect Clinically Actionable Variants. Mol Diagn Ther. 2020;24(3):339-349.
3. Heydt C, Rehker J, Pappesch R, et al. Analysis of tumor mutational burden: correlation of five large gene panels with whole exome sequencing. Sci Rep. 2020;10(1):11387.
4. Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7(3):746-756.
5. Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703-713.
6. Drilon A, Wang L, Arcila ME, et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin Cancer Res. 2015;21(16):3631-363.
7. Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Annals of Oncology. 2015;26(7):1415-1421.
8. Gutierrez ME, Choi K, Lanman RB, et al. Genomic Profiling of Advanced Non-Small Cell Lung Cancer in Community Settings: Gaps and Opportunities. Clin Lung Cancer. 2017;18(6):651-659.
9. Singal G, Miller PG, Agarwala V, et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA. 2019;321(14):1391-1399.
10. Kato S, Kim KH, Lim HJ, et al. Real-world data from a molecular tumor board demonstrate improved outcomes with a precision N-of-One strategy. Nat Commun. 2020;11(1):4965.
11. Sicklick JK, Kato S, Okamura R, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med. 2019;25(5):744-750.

